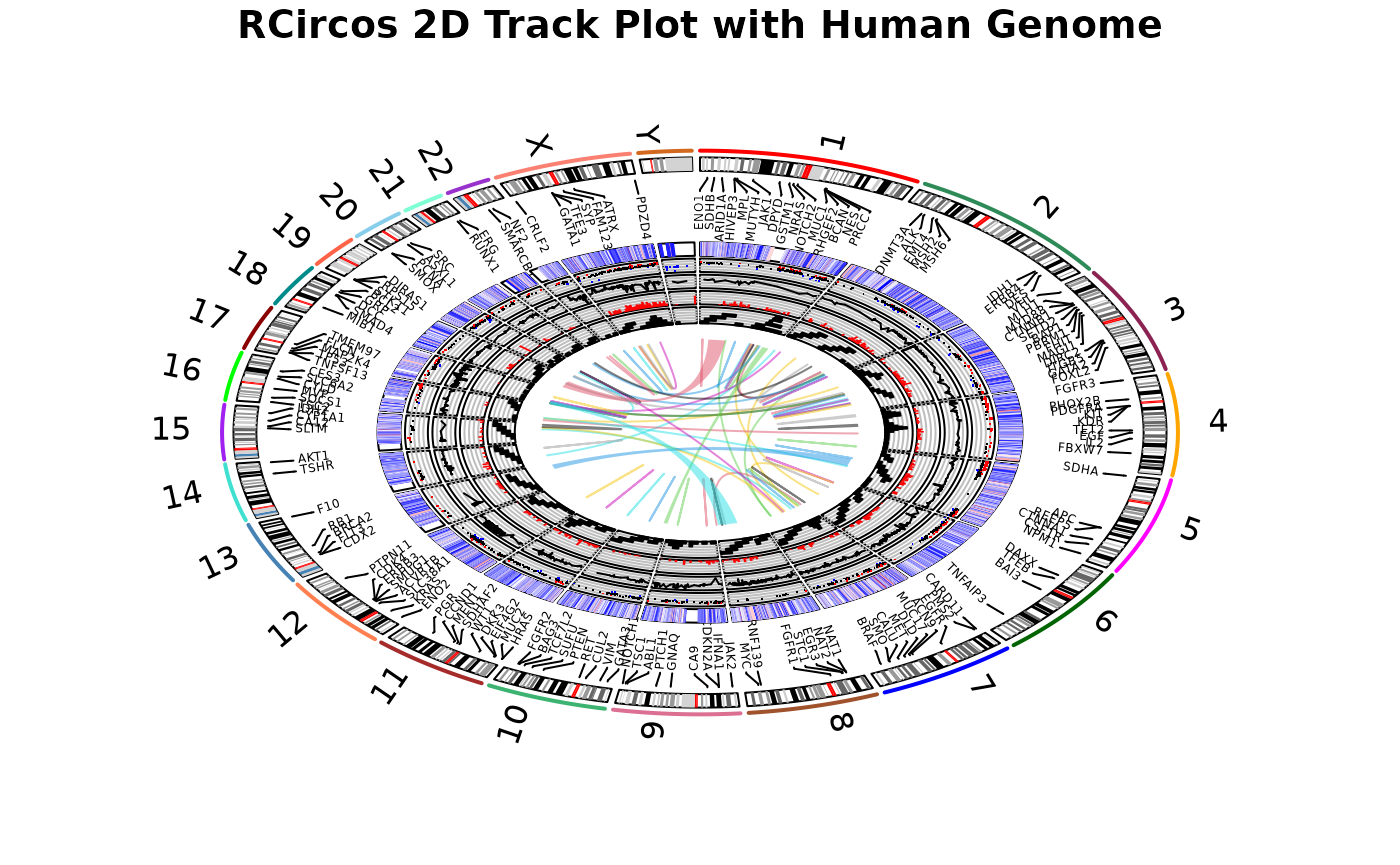
1 RCircos plot with human genomic data
Authors: Hongen Zhang Reviser: Tianze Cao
2025-06-04
Source:vignettes/01 Demo_Human.Rmd
01 Demo_Human.RmdThis demo draw human chromosome ideogram and data tracks for:
- Connectors
- Gene lables
- Heatmap
- Scatterplot
- Line plot
- Histogram
- Tile plot
- Link lines
# Load RCircos package
library(RCircos);
# Load human cytoband data
data(UCSC.HG19.Human.CytoBandIdeogram);
hg19.cyto <- UCSC.HG19.Human.CytoBandIdeogram;
# Setup RCircos core components:
RCircos.Set.Core.Components(cyto.info=hg19.cyto, chr.exclude=NULL, tracks.inside=10, tracks.outside=0);##
## RCircos.Core.Components initialized.
## Type ?RCircos.Reset.Plot.Parameters to see how to modify the core components.
# Open the graphic device (here a pdf file)
message("Open graphic device and start plot ...\n");## Open graphic device and start plot ...
RCircos.Set.Plot.Area();
title("RCircos 2D Track Plot with Human Genome");
# Draw chromosome ideogram
message("Draw chromosome ideogram ...\n");## Draw chromosome ideogram ...
RCircos.Chromosome.Ideogram.Plot();
# Connectors in first track and gene names in the second track.
message("Add Gene and connector tracks ...\n");## Add Gene and connector tracks ...
data(RCircos.Gene.Label.Data);
RCircos.Gene.Connector.Plot(genomic.data=RCircos.Gene.Label.Data, track.num=1, side="in");## Not all labels will be plotted.## Type RCircos.Get.Gene.Name.Plot.Parameters()## to see the number of labels for each chromosome.
RCircos.Gene.Name.Plot(gene.data=RCircos.Gene.Label.Data, name.col=4, track.num=2, side="in");## Not all labels will be plotted.## Type RCircos.Get.Gene.Name.Plot.Parameters()## to see the number of labels for each chromosome.
# Heatmap plot. Since some gene names plotted above are longer than one track height, we skip two tracks
message("Add heatmap track ...\n");## Add heatmap track ...
data(RCircos.Heatmap.Data);
RCircos.Heatmap.Plot(heatmap.data=RCircos.Heatmap.Data, data.col=6, track.num=5, side="in");
# Scatterplot. Note: There is no prefix of "chr" in chromosome names of RCircos.Scatter.Data
message("Add scatterplot track ...\n");## Add scatterplot track ...
data(RCircos.Scatter.Data);
RCircos.Scatter.Plot(scatter.data=RCircos.Scatter.Data, data.col=5, track.num=6, side="in", by.fold=1);
# Line plot. Note: There is no prefix of "chr" in chromosome names of RCircos.Line.Data
message("Add line plot track ...\n");## Add line plot track ...
data(RCircos.Line.Data);
RCircos.Line.Data[,1] <- paste0("chr", RCircos.Line.Data[,1])
RCircos.Line.Plot(line.data=RCircos.Line.Data, data.col=5, track.num=7, side="in");
# Histogram plot
message("Add histogram track ...\n");## Add histogram track ...
data(RCircos.Histogram.Data);
RCircos.Histogram.Plot(hist.data=RCircos.Histogram.Data, data.col=4,
track.num=8, side="in");
# Tile plot. Note: tile plot data have chromosome locations and each data file is for one track
message("Add tile track ...\n");## Add tile track ...
data(RCircos.Tile.Data);
RCircos.Tile.Plot(tile.data=RCircos.Tile.Data, track.num=9, side="in");## Tiles plot may use more than one track. Please select correct area for next track if necessary.
# Link lines. Link data has only paired chromosome locations in each row and link lines are always drawn inside of chromosome ideogram.
message("Add link track ...\n");## Add link track ...
data(RCircos.Link.Data);
RCircos.Link.Plot(link.data=RCircos.Link.Data, track.num=11,
by.chromosome=FALSE);
# Add ribbon link to the center of plot area (link lines).
message("Add ribbons to link track ...\n");## Add ribbons to link track ...
data(RCircos.Ribbon.Data);
RCircos.Ribbon.Plot(ribbon.data=RCircos.Ribbon.Data, track.num=11,
by.chromosome=FALSE, twist=FALSE);