6 RCircos plot showing data track layout
Authors: Hongen Zhang Reviser: Tianze Cao
2025-06-05
Source:vignettes/06 Layout_Demo.Rmd
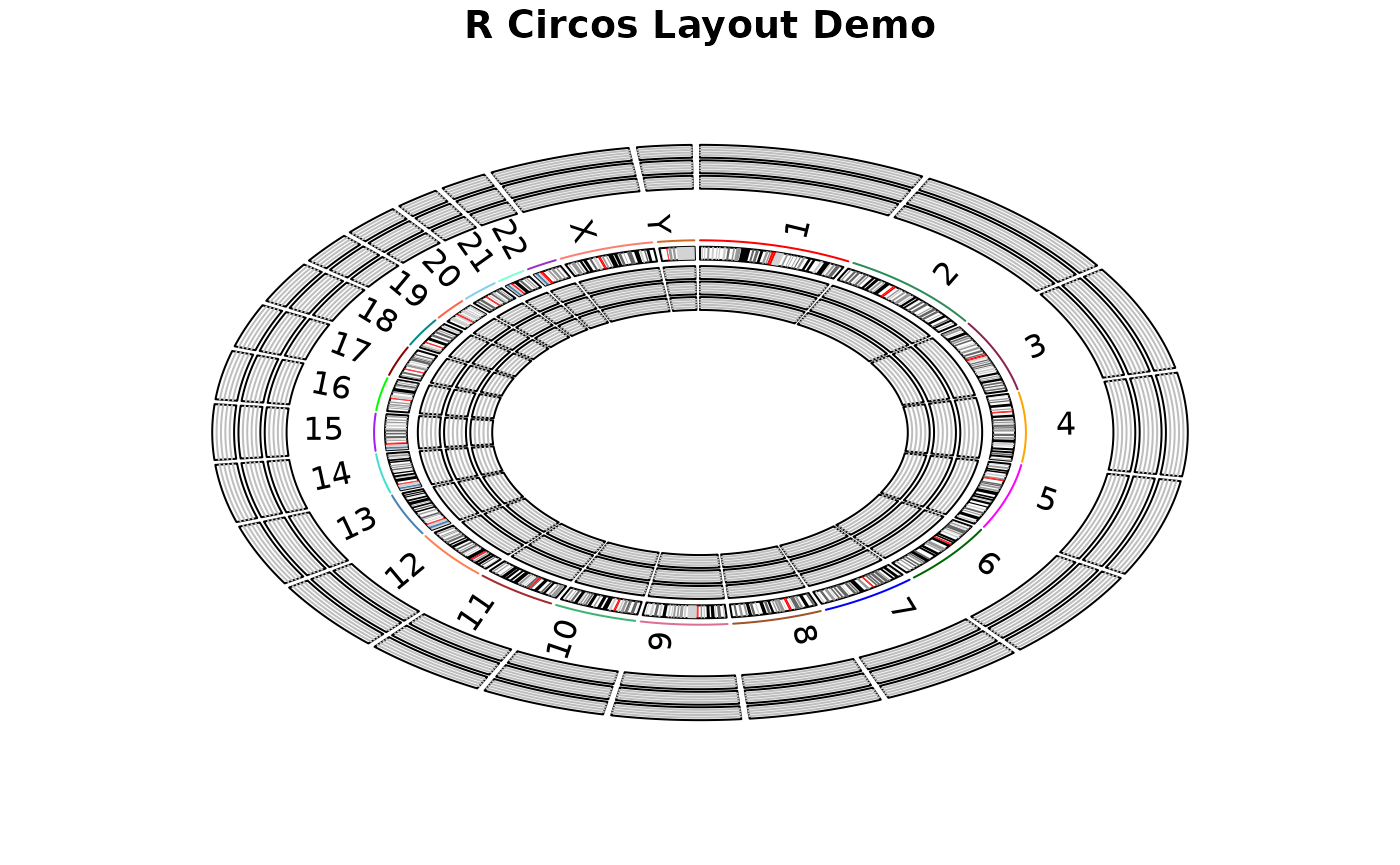
06 Layout_Demo.RmdThis demo draw chromosome ideogram with padding between
chromosomes, highlights, chromosome names, and three empty tracks inside
and outside of chromosome ideogram.
# Load RCircos library
library(RCircos);
# Load human cytoband data
data(UCSC.HG19.Human.CytoBandIdeogram);
cyto.info <- UCSC.HG19.Human.CytoBandIdeogram;
# Setup RCircos core components:
RCircos.Set.Core.Components(cyto.info, NULL, 5, 5);##
## RCircos.Core.Components initialized.
## Type ?RCircos.Reset.Plot.Parameters to see how to modify the core components.
# Open the graphic device (here a pdf file)
RCircos.Set.Plot.Area();
# Draw chromosome ideogram
message("Draw chromosome ideogram ...\n");## Draw chromosome ideogram ...
RCircos.Chromosome.Ideogram.Plot();
title("R Circos Layout Demo");
# Marking plot areas both inside and outside of
# chromosome ideogram ( 3 for each)
# ********************************************
total.track <- 3;
subtrack <- 5;
# Outside of chromosome ideogram
RCircos.Par <- RCircos.Get.Plot.Parameters();
one.track <- RCircos.Par$track.height + RCircos.Par$track.padding;
for(a.track in 1:total.track)
{
in.pos <- RCircos.Par$track.out.start + (a.track-1)*one.track;
out.pos <- in.pos + RCircos.Par$track.height;
RCircos.Track.Outline(out.pos, in.pos, subtrack);
}
# Inside of chromosome ideogram
for(a.track in 1:total.track)
{
out.pos <- RCircos.Par$track.in.start - (a.track-1)*one.track;
in.pos <- out.pos - RCircos.Par$track.height;
RCircos.Track.Outline(out.pos, in.pos, subtrack);
}
message("RCircos Layout Demo Done!");## RCircos Layout Demo Done!